A portrait of Rosalind Franklin with a hidden twist hangs on the wall of the Bill and Melinda Gates Coronary heart for Laptop computer Science and Engineering on the School of Washington in Seattle, US.
The portrait is 5 years earlier and is a black acrylic ink painting of Franklin over a collage of virtually 2000 photos. These images are all snapshots of helpful recollections submitted by most people to Luis Ceze, a professor of laptop computer science and engineering. Nonetheless the true shock lies throughout the medium used to paint Franklin. The acrylic ink incorporates synthetic DNA encoded with all the digital data needed to breed each {{photograph}} throughout the collage. ‘The photographs had been encoded in DNA which was subsequently utilized as ink to create a portrait of Rosalind Franklin, certainly one of many pioneers in DNA evaluation,’ says Karin Strauss, senior principal evaluation supervisor at Microsoft Evaluation in Washington, US.

The idea of storing digital data throughout the pattern of adenines (As), thymines (Ts), cytosines (Cs) and guanines (Gs) in synthetic DNA has been floating spherical for a few years. It gives a further compact and long-lasting numerous to binary code (the strings of zeroes and ones) utilized in typical computing. The ultimate dozen or so years has seen a flurry of sturdy examples of the storage of DNA information in movement. Completely different demonstration initiatives embody storing Shakespeare’s 154 sonnets, part of an audio file of Martin Luther King’s 1963 ‘I’ve a dream’ speech and the first episode of the Netflix sequence Biohackers.
‘This thought-about storing digital information in DNA is not going to be a mainly new concept, nonetheless it is turning into an growing variety of viable,’ says Ceze. A major step forward was the formation of the DNA Information Storage Alliance in 2020. This huge commerce and tutorial collaboration is enabling the occasion of an interoperable storage ecosystem – the place the utilized sciences for each stage of knowledge storage and assortment course of shall be appropriate. This could avoid a repeat of the videotape format battle that seen the incompatible Betamax and VHS system go head-to-head throughout the late Seventies and Nineteen Eighties.
An info decision
The logistics of DNA information storage and retrieval varies between demonstration initiatives nonetheless the precept steps are the an identical. First, the information is encoded proper right into a pattern of nucleotide bases, merely because it’s presently encoded into zeroes and ones. A lot of copies of DNA strands with this pattern of bases are then synthesised throughout the lab. Subsequent, the DNA is saved for a timeframe. To retrieve the information, the pattern of bases throughout the DNA is study using sequencing utilized sciences (that had been initially developed for genomic and medical evaluation features). The DNA might be recovered from Ceze’s portrait of Franklin’s, for example, by scraping some paint off.
Storing data in DNA gives many advantages over current methods, with longevity being essential. Information saved by our ancestors affords us with a window into their world. It is nonetheless potential to see cave work from prehistoric situations, view hieroglyphs carved into rocks, and skim eleventh century books. In distinction, at the moment’s storage mediums normally are usually not designed to closing. The provides degrade fairly quickly and playback know-how is shortly outdated, making it robust to retrieve information larger than a decade or so earlier. What variety of of use nonetheless have the means to entry information saved on vinyl info, cassette tapes, videotapes, floppy disks or zip disks? Most new laptops don’t even have a CD or DVD drive anymore. ‘Museums and large information storage corporations are acutely aware that there is this draw back that we don’t truly know discover ways to retailer the information for a really very long time,’ says Robert Grass, a professor of purposeful provides at ETH Zurich in Switzerland.
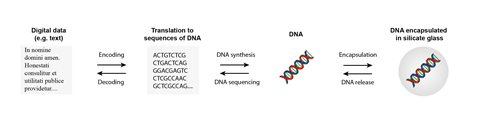
DNA is what nature has developed to utilize to retailer the information that every one residing organisms should develop, reproduce and efficiency. Saved appropriately, it will closing many a whole bunch of years. All through the last few years, scientists have study DNA from one-million-year-old mammoth enamel and from found proof for horseshoe crabs and mastodons in two-million-year-old environmental DNA. ‘DNA lasts a very, very very very long time, notably if it’s saved with out oxygen and water and at midnight,’ says Emily Leproust, chief authorities officer at Twist Bioscience in San Francisco, US.
Moreover it’s terribly probably that future generations will retain the facility to study DNA. ‘DNA is so very important to human nicely being that you just’ll always be succesful to study DNA. In 100 years, maybe we gained’t use Illumina or PacBio anymore, it will be completely totally different sequencing utilized sciences, nonetheless we’ll always be succesful to sequence it,’ Leproust says.
Then there’s the issue of density and vitality consumption. Information saved ‘throughout the cloud’ is unquestionably held in giant information centres scattered all through the globe. The data centres on the Cardiff Information Coronary heart Campus, for instance, have a footprint of about 140,000m2 and devour 270MW of vitality – adequate to help a small metropolis. In opposition to this, ‘DNA is awfully dense,’ says Leproust, ‘you can put dozens of knowledge centres throughout the measurement of a sugar cube.’ It requires no vitality to retailer it and might closing for a whole bunch of years in a sealed container, as long as it is desiccated and saved in a reasonably cool location.
A time capsule
Sooner than DNA information storage can turn into mainstream, two primary hurdles must be overcome: the value of DNA synthesis and sequencing desires to return down and the tempo should go up. Necessary effort is ongoing in course of achieving these goals. It is, nonetheless, unlikely that DNA information storage will ever be low-cost or fast adequate to alternate digital information storage wholesale. In its place, it is anticipated to fill space of curiosity gaps out there out there for features much like archiving information that should be saved for an prolonged timeframe with no must be study pretty typically. Information on this class consists of culturally very important information, licensed paperwork and vital governmental data. ‘I’ve been talking with the Nationwide Archives proper right here throughout the UK and with the British Library,’ says Thomas Heinis, a reader in computing at Imperial College London, UK.
Because of we use a lot much less regents, it’s cheaper
Phosphoramidite chemistry is the predominant methodology used at the moment to synthesise DNA throughout the lab. Synthetic DNA is stitched collectively one nucleotide at a time by forming covalent bonds between the three′ phosphite ester groups and the 5′ hydroxyl groups on adjoining deoxyribose sugar gadgets. Because of the nucleotides are added one after the other and each addition requires security and deprotection steps, setting up synthetic DNA is a laborious and expensive course of.
Miniaturising DNA synthesis is one possibility to chop again costs. DNA is usually made in 96-well plates with one piece of DNA made in each properly. Twist Bioscience has developed an ink-jet printer-based platform that builds 1 million gadgets of DNA concurrently. It makes use of silicon chips (of the type utilized by the semiconductor commerce) that are micropatterned with tiny wells whereby the chemistry takes place. This platform ‘makes use of 99.8% a lot much less chemical substances’ for every little bit of DNA constructed, says Leproust. ‘Because of we use a lot much less regents, it’s cheaper,’ she gives. Twist Bioscience’s know-how is already used to make bespoke synthetic DNA strands for vaccine enchancment, drug discovery, diagnostic design and totally different biotech functions. The early entry of its suppliers for DNA information storage is deliberate for 2025, Leproust says.
An error-filled future
One different methodology getting used to cut synthesis costs (and likewise tempo up sequencing) is utilizing error correction codes. These additional gadgets of DNA proper for any errors so that the information can nonetheless be study. Information strings in digital storage utilized sciences moreover embrace redundant code, which will be utilized to proper any errors if one factor goes incorrect. Being able to proper errors throughout the information study out on the end of the tactic opens the door to a lot much less right nonetheless cheaper and faster synthesis and sequencing devices getting used. ‘We’re capable of do points on the encoding stage, throughout the DNA data itself, to take care of errors,’ explains Jeff Nivala, an assistant professor of laptop computer science and engineering on the School of Washington. ‘I can then deal with a very extreme error cost with my [synthesis or] sequencing gadget as a result of it’s simple for me to proper for that.’ Error correction codes may take care of error launched all through storage. CDs with mild scratches on their flooring can nonetheless be carried out, for example, resulting from error correction codes.
Lower-accuracy DNA synthesis methods being explored for information storage embody massively parallel light-directed synthesis with added error correction codes. Grass and collaborators along with Mark Somoza, a professor of chemistry on the School of Vienna in Austria, are pioneering this methodology. ‘We’re capable of synthesise roughly 2 million sequences in parallel,’ Somoza explains. The strategy removes the protecting groups on the 5′ hydroxyl using UV mild in a motion cell system into which the obligatory reagents for each step are added cyclically. ‘There’s [normally] an acid label defending group on the 5’ website online and we modified that with a photolabel group,’ says Somoza. All through the deprotection, an array of micromirrors is used to direct UV mild very precisely onto DNA’s flooring. The rest of the chemistry used is much like typical DNA synthesis approaches. Gentle-directed synthesis is significantly cheaper and faster than customary DNA synthesis. Using this methodology, the employees has demonstrated flawless information restoration of a file containing sheet music by Mozart.
AI to the rescue
Olgica Milenkovic, a professor of knowledge processing on the School of Illinois at Urbana–Champaign throughout the US is exploring one different methodology to coping with errors: artificial intelligence (AI). ‘Synthetic DNA is so expensive [that] using error correction coding may add an enormous overhead,’ she explains. ‘We use the gathering of [already developed] methods for machine learning and artificial intelligence to make the images encoded in DNA look larger throughout the presence of errors, pretty than to try to restore the errors.’ This methodology isn’t applicable for information that should be extraordinarily right, nonetheless it does work properly for images the place AI devices exist already to ‘restore’ damages to earlier photos so that they’re not seen to the naked eye.
Phosphoramidite chemistry could also be very dirty, very toxic, very laborious and expensive
Milenkovic has moreover developed a definite writing methodology with the information saved throughout the DNA in two strategies. The image data is positioned in nucleotide patterns throughout the synthetic DNA using typical DNA synthesis methods. Copyright particulars and watermarks are then added as a pattern of nicks throughout the DNA backbone. These are a binary code and made by nicking enzymes. ‘When you may have a nick it means it’s a one, if you’ve received no nick it means it’s a zero,’ Milenkovic explains. Using two layers of coding means further data might be saved within the an identical home. Milenkovic and her group have used their methodology to retailer and reproduce eight Marlon Brando movie stills.
Enzymatic synthesis will also be being explored for setting up synthetic DNA. This know-how is far much less properly developed than phosphoramidite chemistry nonetheless has the potential to supply a faster and cheaper numerous that doesn’t require toxic chemical substances. ‘Phosphoramidite chemistry could also be very dirty, very toxic, very laborious and expensive,’ says Ceze. ‘We’ve got to truly dwelling in on enzymatic synthesis [to make it] controllable and extreme throughput as properly.’ Kern Strategies, a spin-out from George Church’s Harvard lab, and France-based DNA Script are among the many many corporations that are advancing enzymatic synthesis of DNA for information storage.
Tempo learning
Sequencing by synthesis methods much like Illumina’s sequencing platforms are the current gold commonplace for learning once more information saved in DNA. Nanopore sequencing is gaining traction due to its talent to sequence single molecules of DNA with out the need for amplification. These models use molecular motors to ratchet DNA strands by means of a pore in a polymer membrane that comes with a detector. The ions throughout the surrounding decision motion by means of the pore producing {an electrical} current and as each base passes by means of the pore it creates a definite (measurable) distortion of this current. The know-how is ‘a really good choice to [achieve] extraordinarily right sequencing’, explains Nivala.
As soon as we do a DNA information storage experiment in the meanwhile, we might like numerous PhD school college students pipetting stuff
Neither sequencing by synthesis or at the moment’s nanopore sequencing models are fast adequate for DNA information storage functions. Oxford Nanopore’s industrial models, for example, have a most tempo of 400 bases per second. Work to develop faster and cheaper nanopore sequencers is ongoing in tutorial and industrial labs throughout the globe. ‘If we are going to do away with these molecular motors and use electrophoretic vitality or the voltage vitality all through the gadget itself, you can push these DNA strands by means of these nanopores orders of magnitude faster,’ Nivala gives. Compromising sequencing accuracy will even allow the worth per gigabyte of knowledge retrieved to go down. For context, Oxford Nanopore’s disposable dongle-sized capsules (the Flongle) worth $90 (£71) each and may sequence as a lot as 2.6Gb of knowledge in 16 hours. That’s regarding the amount of knowledge needed to retailer a film like Star Wars: The Ultimate Jedi in commonplace definition.
Automation of your complete storage course of is one different area of focus. The synthesis and sequencing utilized sciences for the time being are principally automated, nonetheless the intermediate steps are nonetheless largely handbook. ‘As soon as we do a DNA information storage experiment, I’ve numerous PhD school college students shifting throughout the lab pipetting stuff,’ Grass explains.
It’s going to be essential to utterly automate the write-to-store-to-read cycle for DNA information storage to vary into mainstream and be viable for functions previous archival information that is hardly accessed, wrote Strauss and Ceze in an article in 2019 that outlined an automated end-to-end occasion of DNA information storage. Their benchtop-size organize first converts the information (with additional error correction code) from zeroes and ones into As, Ts, Cs and Gs. These bases are then pumped in order onto a column the place they’re stitched collectively using phosphoramidite chemistry. As quickly because the strands are full, they’re washed off the secure helps on the column and proper right into a storage bottle. To retrieve the information, the liquid is pumped into an Oxford Nanopore’s MinION gadget the place the DNA is sequenced. Lastly, this code of As, Ts, Cs and Gs is decoded once more into zeroes and ones. Of their first demonstration, the scientists despatched the phrase ‘good day’ in 21 hours. ‘Although we have now now demonstrated that it is potential to utterly automate an end-to-end DNA information storage system at low worth, that system was not extreme throughput,’ says Strauss. Work is ongoing to scale up and tempo up this automated methodology.
There’s clearly nonetheless rather a lot work to be carried out sooner than DNA information storage turns into mainstream. These working throughout the self-discipline think about that it’s solely a matter of time sooner than just a few of our giant vitality hungry information centres will start to get changed by tiny capsules of DNA that our ancestors in a whole bunch of years’ time will probably be succesful to entry. And that poses a question: what message would you most prefer to go away individuals who observe in your footsteps?
Nina Notman is a science writer based in Salisbury, UK
